Accurately counting the number of cells in microscopy images is required in many medical diagnosis and biological studies. This task is tedious, time-consuming, and prone to subjective errors. However, designing automatic counting methods remains challenging due to low image contrast, complex background, large variance in cell shapes and counts, and significant cell occlusions in two-dimensional microscopy images. In this study, we proposed a new density regression-based method for automatically counting cells in microscopy images.
Methodology:
As shown in the figure, the proposed fromework integrates a concatenated neural network design and deeply-supervised learning strategy to learn an end-to-end mapping from a given image to the density map of cells. The number of cells is computed by summing the estimated density map up.

Results:
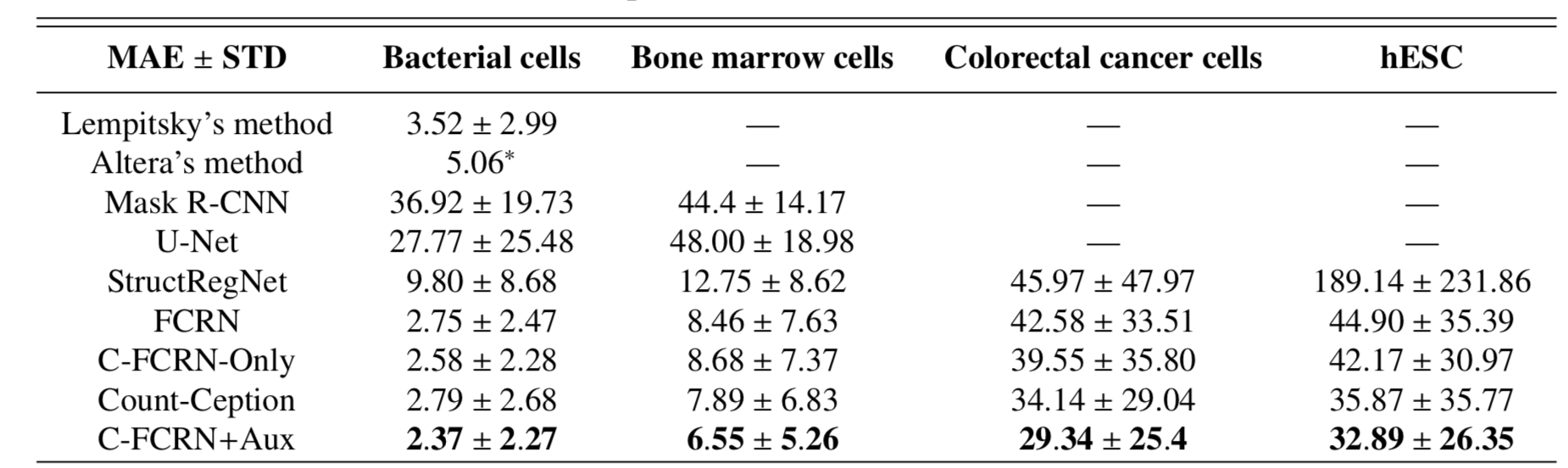
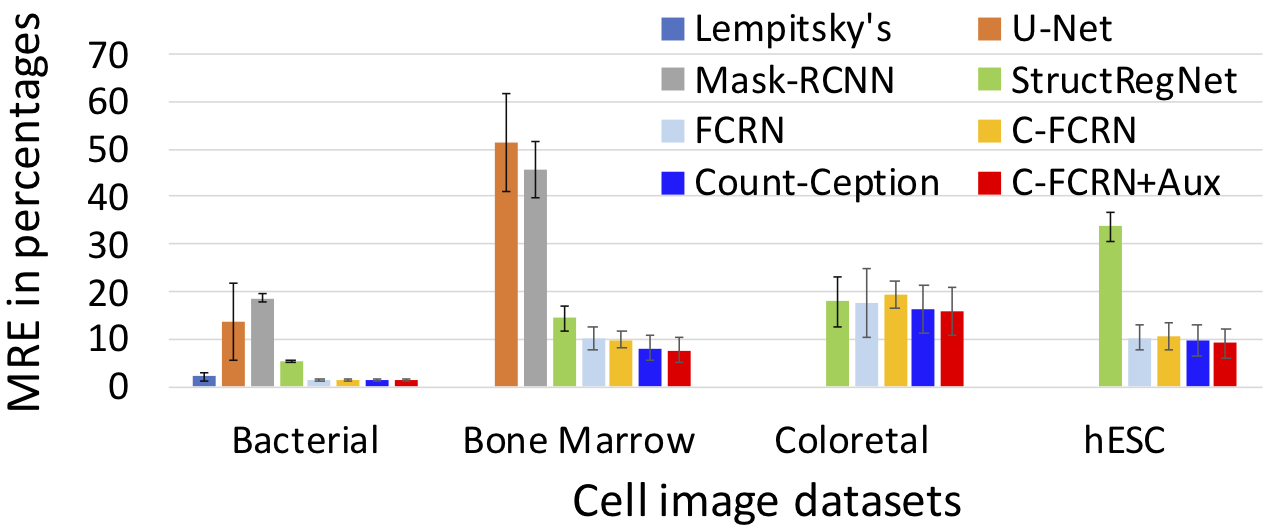
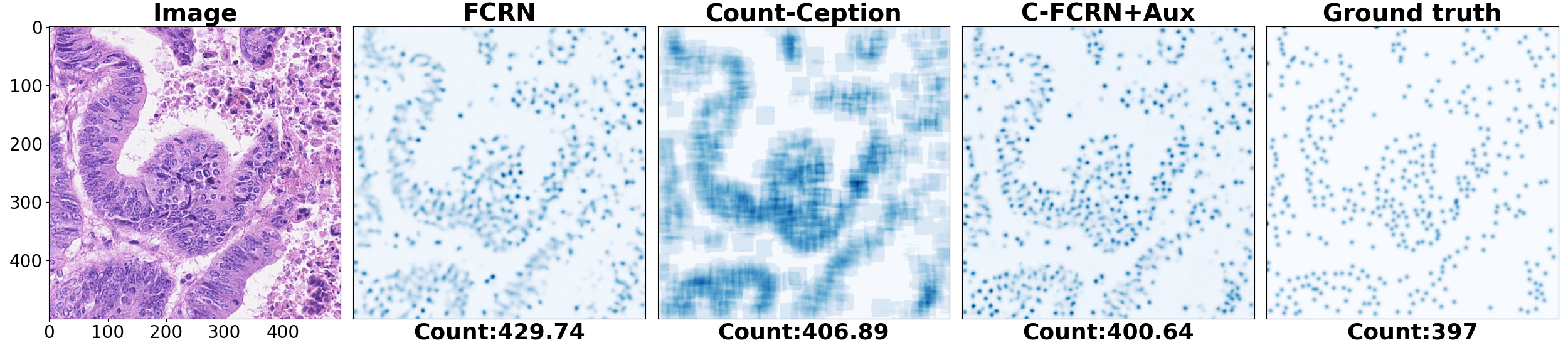
To quantitively evaluate our proposed method, mean absolute count error (MAE) and mean relative count error (MRE) related to our method was analyzed. 5-fold cross validation was used for the evaluation. The method was tested on four challenging datasets separately. The performance of our method was compared to 8 state-of-the-art methods, including density regression, count regression and cell detection based counting methods. The corresponding results are shown in the Figure 2 and Figure 3. The prediction results for an example colorectal cancer cell image is shown in Figure 4.
Related publications:
1. Shenghua He, Kyaw Thu Minn, Lilianna Solnica-Krezel, Mark A. Anastasio, and Hua Li*, “Deeply-supervised density regression for automatic cell counting in microscopy images”, Medical Image Analysis 68 (2020): 101892.
2. Shenghua He, Kyaw Thu Minn, Lilianna Solnica-Krezel, Hua Li, and Mark Anastasio, “Automatic microscopic cell counting by use of unsupervised adversarial domain adaptation and supervised density regression”, SPIE Medical Imaging 2019.
3. Shenghua He, Kyaw Thu Minn, Lilianna Solnica-Krezel, Mark Anastasio, and Hua Li*, “Automatic microscopic cell counting by use of deeply-supervised density regression model,” SPIE Medical Imaging 2019.